Plant Disease Detection with Keras and FastAPI
Plant Disease Detection model built with Tensorflow and deployed as an API on Heroku with FastAPI. An end-to-end Machine Learning Project carried out by Group 1 Zummit Africa AI/ML Team to detect disease (Rust, Powdery Mildew) of infected plants.
You can check out the article on Hashnode describing in detail how this project was carried out.
The App can be can be viewed here
Contributors
Table of Contents
- Plant Disease Detection with Keras and FastAPI
Repository File Structure
├── app.py # main fastapi app
├── aws.txt # requirements for deploying on aws
├── docker-compose.yml # for running docker compose
├── Dockerfile
├── download.py # script to download model hosted on google drive
├── fastapi_setup # for deploying on aws
├── LICENSE
├── Notebook #Notebook folder
└── plant_disease_detection.ipynb
├── README.md
├── requirements.txt
├── Research_Papers
│ └── R_paper1.pdf
├── setup.sh # bash script for deploying on aws ec2
└── test_images # test images for inference
├── 8fd27998ae52a4a6.jpg
├── 9be41b823d13e3c6.jpg
└── 9d0f6e60819f9a5a.jpg
Problem Statement
In this project, a neural network model was built using Tensorflow. The model detects if a plant is suffering from a disease(Rust or Powdery Mildew). The model was then deployed as an API using the FastAPI framework.
Rusts
Rusts are plant diseases caused by pathogenic fungi of the order Pucciniales (previously known as Uredinales). An estimated 168 rust genera and approximately 7,000 species, more than half of which belong to the genus Puccinia, are currently accepted. Rust fungi are highly specialized plant pathogens with several unique features. Taken as a group, rust fungi are diverse and affect many kinds of plants. However, each species has a very narrow range of hosts and cannot be transmitted to non-host plants. In addition, most rust fungi cannot be grown easily in pure culture. A single species of rust fungi may be able to infect two different plant hosts in different stages of its life cycle, and may produce up to five morphologically and cytologically distinct spore-producing structures viz., spermogonia, aecia, uredinia, telia, and basidia in successive stages of reproduction. Each spore type is very host specific, and can typically infect only one kind of plant. Rust fungi are obligate plant pathogens that only infect living plants. Infections begin when a spore lands on the plant surface, germinates, and invades its host. Infection is limited to plant parts such as leaves, petioles, tender shoots, stem, fruits, etc. Plants with severe rust infection may appear stunted, chlorotic (yellowed), or may display signs of infection such as rust fruiting bodies. Rust fungi grow intracellularly, and make spore-producing fruiting bodies within or, more often, on the surfaces of affected plant parts.

Powdery Mildew
Powdery mildew is a fungal disease that affects a wide range of plants. Powdery mildew diseases are caused by many different species of fungi in the order Erysiphales. Powdery mildew is one of the easier plant diseases to identify, as its symptoms are quite distinctive. Infected plants display white powdery spots on the leaves and stems. The lower leaves are the most affected, but the mildew can appear on any above-ground part of the plant. As the disease progresses, the spots get larger and denser as large numbers of asexual spores are formed, and the mildew may spread up and down the length of the plant. Powdery mildew grows well in environments with high humidity and moderate temperatures. Greenhouses provide an ideal moist, temperate environment for the spread of the disease. This causes harm to agricultural and horticultural practices where powdery mildew may thrive in a greenhouse setting. In an agricultural or horticultural setting, the pathogen can be controlled using chemical methods, bio organic methods, and genetic resistance. It is important to be aware of powdery mildew and its management as the resulting disease can significantly reduce important crop yields.

Project Outline
- Analyse the image data.
- Build a Neural Network with Keras for classifying the images.
- Design, build and deploy the model as a FastAPI Application.
Data Preparation
The dataset used to train the neural network contains three labels, **“Healthy”, “Powdery”, “Rust” referring to plant conditions. There is a total of 1530 images divided into train, test, and validation sets.**
Model Building
The model was built using Keras(Tensorflow as backend) and was trained using the train images in the dataset and then evaluated using the test images in the dataset. The model was trained using the following parameters:
- nb_train_samples = 400
- nb_validation_samples = 100
- nb_eval_samples = 60
- epochs = 50
- batch_size = 16
- num_of_class = 3
After training, the model had an accuracy of 93.75%
Model Diagram

Model Accuracy

Model Loss

Preview
FastAPI Demo
https://user-images.githubusercontent.com/101701760/181192761-a3c4e9d7-5519-43d8-9a27-f9ca7896cce8.mp4
How to run the Application
Running on Local Machine
To run the application on your local system do the following:
- Clone the repository:
git clone https://github.com/Nneji123/Plant-Disease-Detection-Keras.git - Change the directory:
cd Plant-Disease-Detection-Keras - Install the requirements:
pip install -r requirements.txt - Download the model from google drive
python download.py - Run the application
uvicorn app:app --reload --port 8000You should be able to view the application by going to http://127.0.0.1:8000/
Running on Local Machine with Docker Compose
You can also run the application in a docker container using docker compose(if you have it installed)
- Clone the repository:
git clone https://github.com/Nneji123/Plant-Disease-Detection-Keras.git - Change the directory:
cd Plant-Disease-Detection-Keras - Download the model:
pip install gdown python download.py - Run the docker compose command
docker compose up -d --buildYou should be able to view the application by going to http://localhost:8000/
Running in a Gitpod Cloud Environment
Click the button below to start a new development environment:
Deployment
Deploying the Application to Heroku
Assuming you have git and heroku cli installed just carry out the following steps:
- Clone the repository:
git clone https://github.com/Nneji123/Plant-Disease-Detection-Keras.git - Change the directory:
cd Plant-Disease-Detection-Keras - Login to Heroku
heroku login
heroku container:login
- Create your application
heroku create your-app-nameReplace your-app-name with the name of your choosing.
- Build the image and push to Container Registry:
heroku container:push web
- Then release the image to your app:
heroku container:release web
How to deploy the application on AWS EC2 using a Bash Script
1. Fork this repository
2. Login to AWS, create a new AWS EC2 instance and make sure to allow outside traffic as shown in the screenshots below:
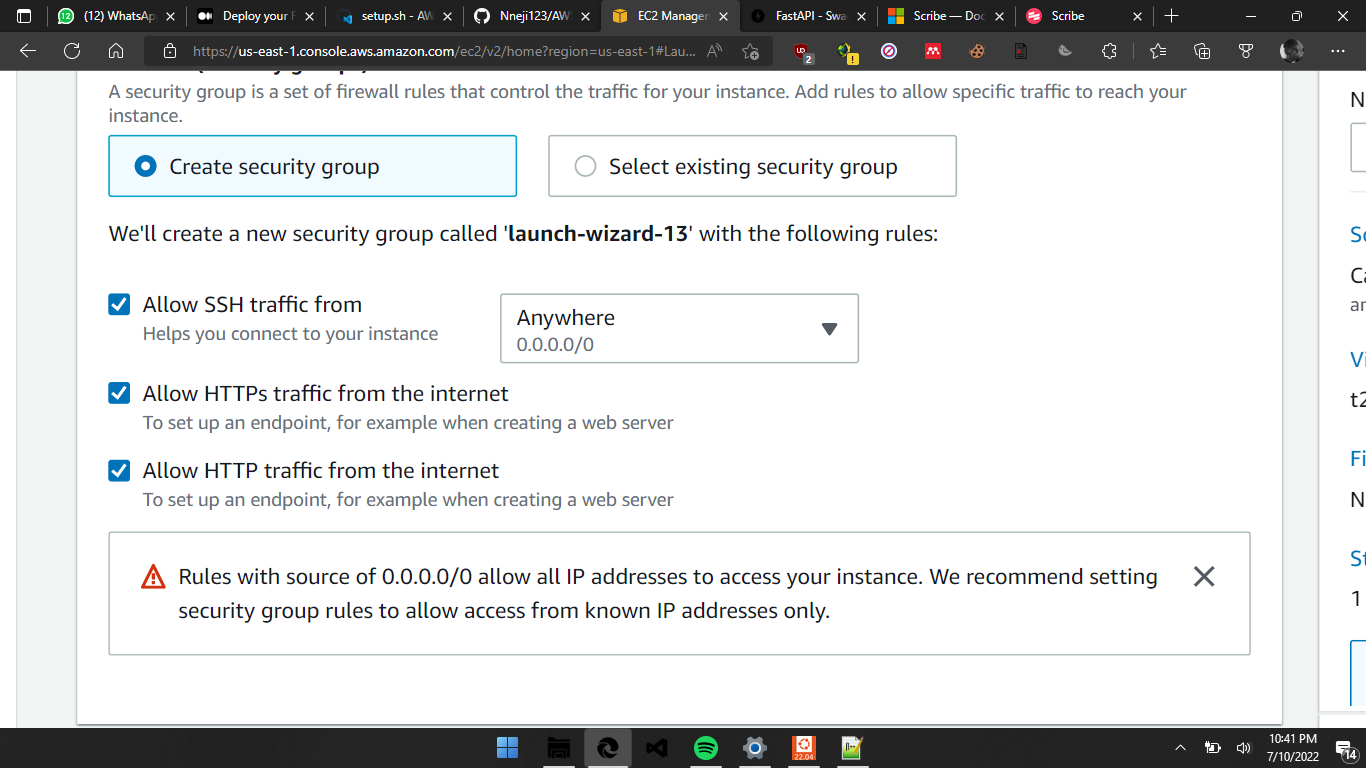
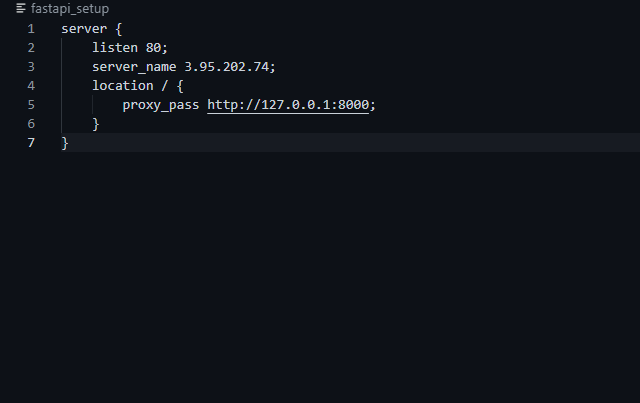
3. When the instance has been launched, copy the Public IP address of your instance and paste it in the ‘fastapi_setup’ file of your cloned repository as shown below
4. Connect to your instance and clone your forked repository, an example in my case:
git clone https://github.com/Nneji123/Plant-Disease-Detection-Keras.git
5. cd into your repository which is probably named ‘Plant-Disease-Detection-Keras’. You can do that by running:
cd Plant-Disease-Detection-Keras
6. Then run the setup.sh file to get your application up and running:
chmod u+x setup.sh
./setup.sh
You can then view the application by going to your Public IP’s location, an example in my case will be: http://3.95.202.74:80/docs
You can also watch this video for a more in depth explanation on how to deploy a FastAPI application on AWS EC2:







